Cell Line Authentication with Deep Sequencing
Patented Technology: Ensuring Precise and Reliable Biosample Authentication
Are your cell lines and biological models REALLY what you think they are? According to multiple studies, 1/5 to 1/3 of popular cell lines have been contaminated by intraspecies/interspecies cells, mycoplasma and/or viruses. The International Cell Line Authentication Committee (ICLAC) has documented a growing list of more than 530 misidentified cell lines (update in Jan 2023) having no known authentic stock.
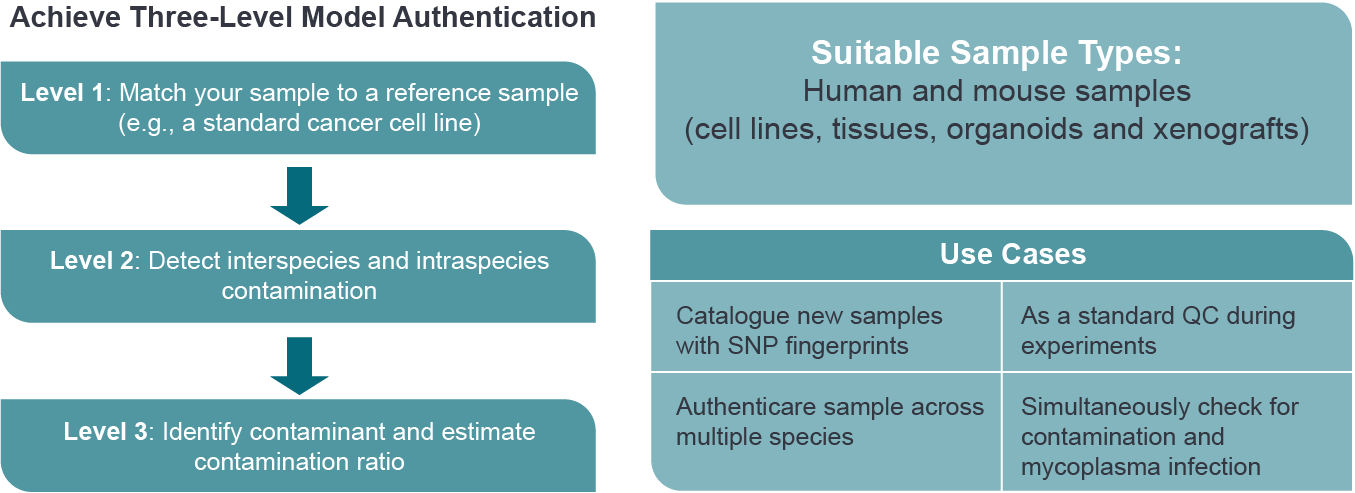
Ensure confidence in your models and reproducibility of high quality data by using our deep sequencing-based cell line authentication service. The panel encompasses over 2000 SNPs and chromosome segments to accurately characterize mouse and human samples including cell lines, organoids, xenograft models, and patient tissues. We have also generated unique DNA fingerprints for the most commonly used oncology research models, covering over 1,200 human cancer and 90 mouse cell lines.
Conventional PCR-based STR assays target only 9 to 24 STR sites, depending on the vendors. The accuracy of the PCR-based STR assay can be low in cases of close genetic relationships such as the authentication of murine cell lines derived from specific strains of inbred animals that lack unique genetic markers and different tumor cell lineages from the same human donor. Further, biosamples with mutations in mismatch repair (MMR) genes are known to exhibit microsatellite instability and a hypermutator phenotype. Consequently, this can lead to genetic drift and/or outgrowth of contaminating cells and STR misclassification.
Cell Line Authentication
with Deep Sequencing

With deep sequencing-based QC panel, you will achieve three-level sample authentication.
"We have used STR assays for cell line authentication in the past, but found that this technique lacks the sensitivity and ability to detect mouse cell contamination. Unlike other services, the cell line authentication service from Crown Bioscience provides a high-level of sensitivity and accuracy, and the ability to detect interspecies, and other, contamination so we can continue our ground-breaking research."
Professor Hongchun Liu,
Shanghai Institute of Materia Medica, Chinese Academy of Sciences
| Cell Line (CLA) Authentication Assay Comparison |
CLA with Deep Sequencing | CLA with PCR-based STR Assay |
|---|---|---|
| Technology | Barcode deep sequencing |
|
| Readout Type | Digital (clean, near-zero quantification error) |
Analog (noisy, high quantification error) |
| Human Cell Authentication | Yes | Yes |
| Mouse Cell Authentication | Yes | Limited |
| MMR Deficient Cell lines identification | Yes | No |
| Contamination-Detecting Sensitivity | High (1%) | Low to medium (5-20%) |
| Accuracy | High | Low to medium |
| Throughput | High | Low |
| Contaminant Identification | Yes | No |
| Qualification of Contamination Ratio | Yes | No |
| Suitable for Large Biobanks | Yes | No |
| Interspecies Contamination Detection | Yes | Limited |
| Intraspecies Contamination Detection | Yes | Limited |
| Detecting Contamination w/o Reference | Yes | No |
| Estimating Mix Ratios for 3+ Cell Lines | Yes (1% sensitivity) | No |
List of authenticatable human and mouse cell lines
NAR Genomics and Bioinformatics, Volume 2, Issue 3, September 2020, lqaa060
Sample Requirements and Submission
| Sample Type | Amount | Shipping Conditions | |
|---|---|---|---|
| DNA | Powder | 100-500μg | Room tempature (RT) |
| Liquid | Wet ice | ||
| Cell Pellet | At least 1 million cells |
Dry ice |
|
| Fresh Tumor Tissue | Snap Frozen | At least 200mg or 1 million cells | Dry ice |
| Formalin Fixed | RT for delivery within 72hr | ||
| FFPE Tissue | Unstained tissue sections (and H&E stained adjacent sections) | RT | |
FAQs
SNP is especially useful for cells:
- With deficiencies in mismatch repair/microsatellite instability
- Derived from closely related inbred strains
- With chromosomal copy number aberrations
7-11 business days
Human gender is inferred via 3 SNPs on the Y chromosome. Please note that the Y chromosome is lost in some cancer genomes, therefore a predicted result of ‘female’ may not always be accurate.
Our panel can classify three reference populations, Han Chinese (CHB), Nigeria Yoruba (YRI), and Utah residents with Northern and Western European ancestry (CEU). The race can be predicted when >70% of total SNPs support one population.
Greater than 90% similarity ratio is considered a match.