- Our Services
- Platforms
- Target Solutions
- Technologies
- Service Types
- Our Science
- About Us
- Contact us

We offer high-throughput sequencing using second and third generation sequencing and optical genome mapping platforms to deliver high-quality, interpretable results.
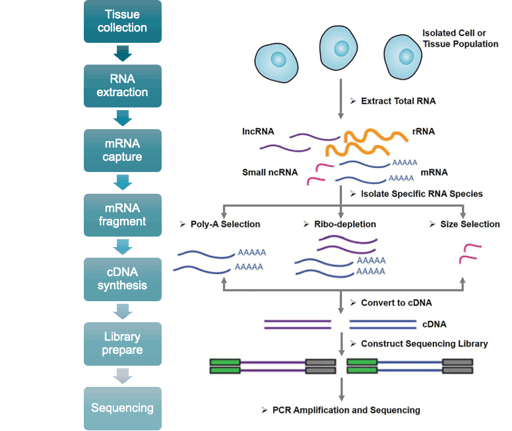
Our RNA-Seq platforms are compatible with a variety of sample types, including cell lines, snap-frozen tissue, whole peripheral blood, FFPE blocks and sections, peripheral blood mononuclear cells (PBMCs), total RNA, and biopsy tissue. Compatible library preparation kits include non-Illumina for mRNA, Illumina Truseq for mRNA, and ribosomal RNA-seq for mRNA, lncRNA, and microRNA.
Applications:
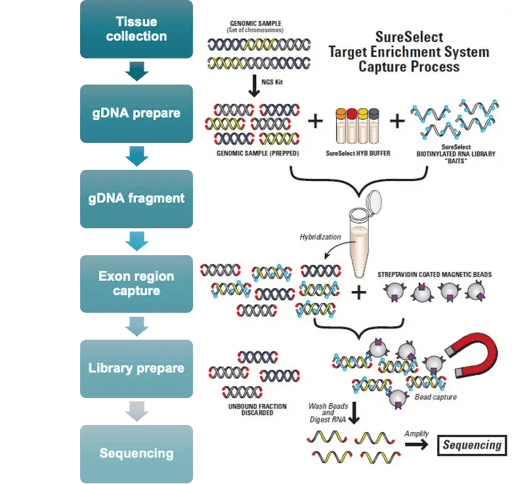
Our WES service is suitable for sequencing-based analysis of cell lines, snap-frozen tissue, whole peripheral blood, FFPE blocks and sections, PBMCs, biopsy tissue, and mouth swabs.
Applications:
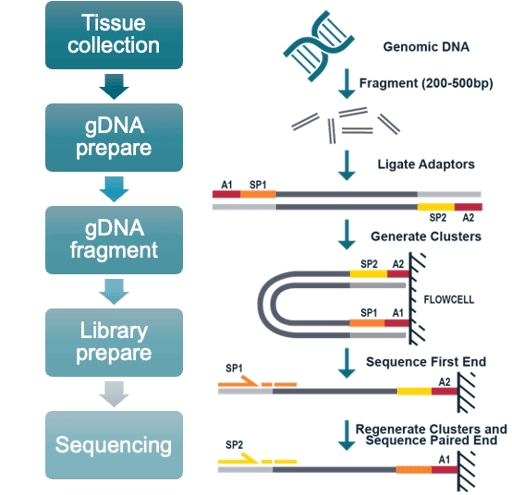
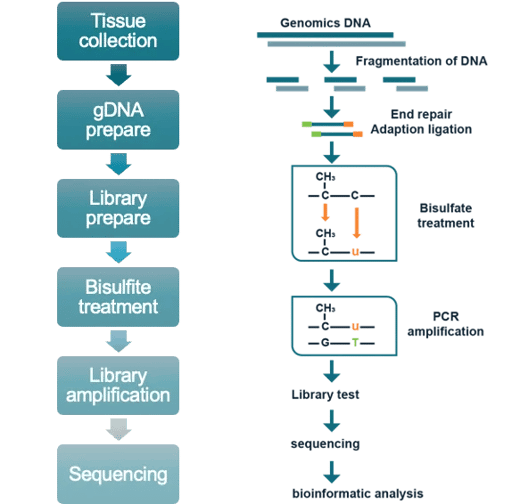
WGS/WGBS platforms are compatible with multiple sample types, including cell lines, snap-frozen tissue, whole peripheral blood, FFPE blocks and sections, PBMCs, biopsy tissue, and mouth swabs.
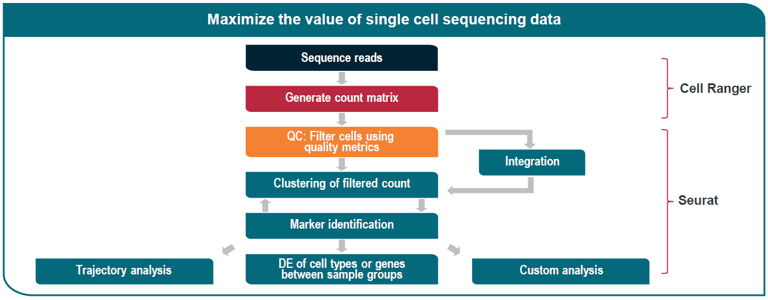
Our single cell sequencing service is suitable for analysis of cell lines, fresh tissue, whole peripheral blood, and PBMCs in single cell RNA-Seq and T cell receptor (TCR) sequencing assays
We also offer bioinformatics analysis for our single cell sequencing service, which includes:
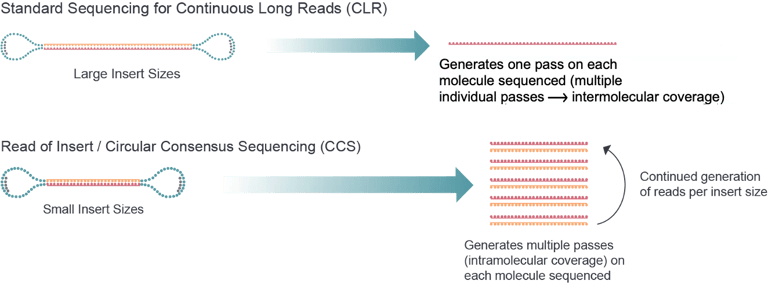
Crown Bioscience uses the PacBio Sequel II platform, which offers single-molecule, real-time (SMRT) sequencing. Long reads are readily assembled into complete genomes for sequencing of full-length transcripts with >99.999% consensus accuracy. Other benefits include uniform coverage of typically inaccessible regions, single-molecule resolution, and direct detection of epigenetic modifications.
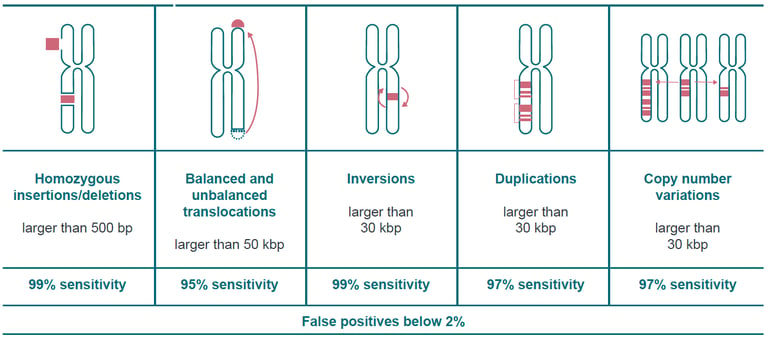
Large structural variations are responsible for many diseases and conditions, including cancers and developmental disorders. Optical genome mapping with Saphyr detects structural variations ranging from 500 bp to megabase pairs in length and offers assembly and discovery algorithms that far outperform sequencing-based technologies in sensitivity.
| Method | Resolution | Features |
|---|---|---|
| Karyotyping | -5-10 Mbp | Extensive training required for interpretation, slow, labor-intensive data collection and analysis. Cell culture required. |
| FISH | -100 kbp | Targeted, extremely limited approach; only shows handful of variants; slow, labor-intensive data collection. Requires validation of every lot of probes utilized |
| Array based techniques |
-50 kbp | Cannot detect balanced rearrangements. Cannot resolve nature of a structural aberration. |
| OGM with Bionano Saphyr |
500 bp | Fully automated detection of: CNVs, repeat expansion, FSHD, unbalanced events from single exon level to aneuploidies, balanced events, inversions, translocations, gene fusions down to 1% VAF. |
We offer advanced microbiome sequencing services, including full-length 16S rRNA analysis, shotgun metagenomic sequencing, and metatranscriptomic sequencing, to provide a comprehensive understanding of the microbiome through genomic sequencing and analysis.
At Crown Bioscience, we offer cutting-edge NanoString nCounter platform services to analyze preclinical and research use only (RUO) clinical samples.
© 2024 Crown Bioscience. All Rights Reserved.


© 2024 Crown Bioscience. All Rights Reserved. Privacy Policy
2024-06-21
2023-03-07
site_page
Genomics