- Our Services
- Platforms
- Target Solutions
- Technologies
- Service Types
- Our Science
- About Us
- Contact us
Microbiome study is vital for drug discovery as the microbiome significantly influences drug metabolism, efficacy, and safety. Understanding these interactions can optimize drug dosages, enhance treatment effectiveness, and enable personalized medicine based on an individual's unique microbiome profile. Moreover, the microbiome's impact on drug-induced toxicity and interactions between medications can be identified, leading to safer drug development. Microbiome-targeted therapies offer promising approaches, directly treating conditions or improving drug effects. Microbiome analysis also helps identify biomarkers for diseases and treatment response, advancing diagnostic tools and precision medicine. Integrating microbiome data into drug development can revolutionize healthcare by improving treatment outcome
Microbiome sequencing provids insights into complex microbial communities. Choose our microbiome sequencing services for:
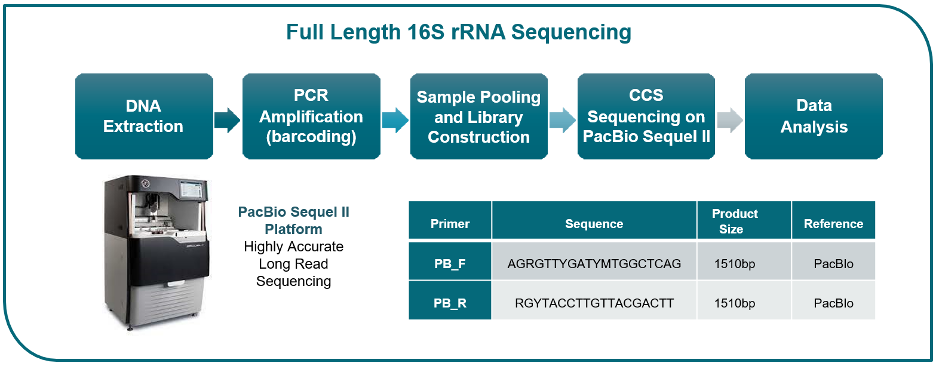
Crown Bioscience leads in comprehensive and cost-effective full-length 16S rRNA analysis using PacBio's long-read third-generation sequencing. Our approach offers distinct advantages over short read sequencing. By capturing the entire 16S rRNA gene, we provide unparalleled insights into bacterial diversity and ensure precise taxonomic classification. Overcoming limitations of short read methods, such as primer bias and reduced taxonomic resolution, our full-length sequencing delivers a comprehensive and unbiased view of the microbial community. This empowers the identification of potential drug targets and deepens understanding of their functional roles.
|
Traditional 16S Sequencing |
Full-Length 16S Sequencing |
|
|
Sequencing Platform |
Short Read Sequencing |
Long Read Sequencing |
|
Targeted Region |
Short hypervariable region |
Entire 16S rRNA gene |
|
Taxonomic Resolution |
Moderate |
Higher |
|
Primer Bias |
Yes |
Reduced |
|
Secondary Structure |
Not captured |
Captured |
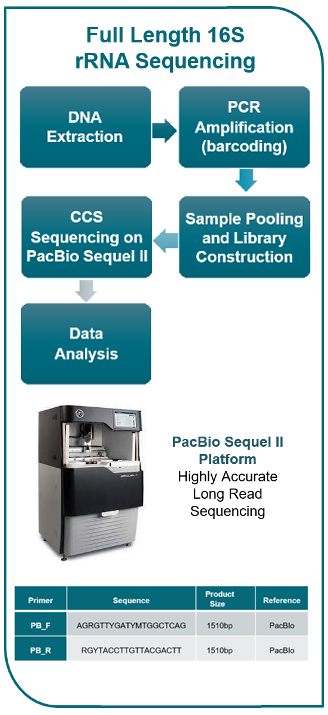
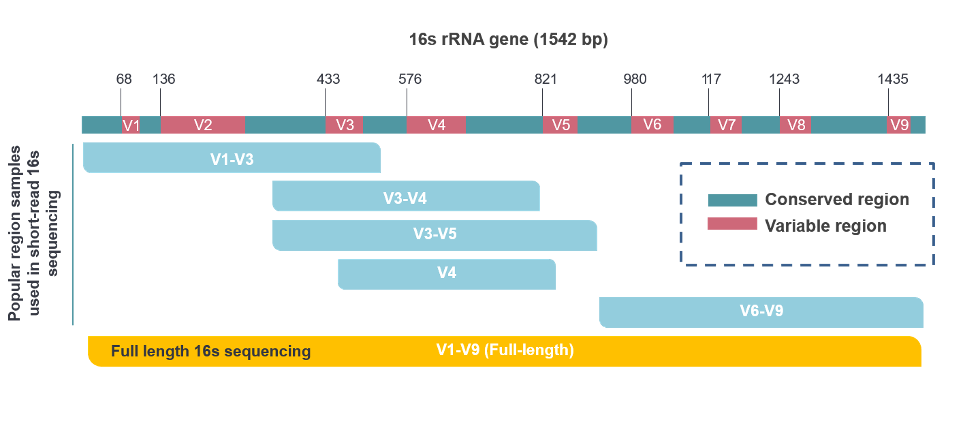

Fig. Commonly used DNA regions in short-read and long-read 16S rRNA sequencing
Metagenomic sequencing provides a significant advantage over 16S rRNA sequencing by capturing comprehensive genomic information. This empowers higher taxonomic resolution, direct functional profiling, and the identification of novel or rare species. While it comes at a higher cost and requires specialized bioinformatics expertise, shotgun sequencing is an ideal choice for researchers looking to delve into a smaller number of samples with greater depth compared to amplicon sequencing. Embrace the power of metagenomic sequencing to unlock a wealth of valuable insights about microbial communities and their functional potential.
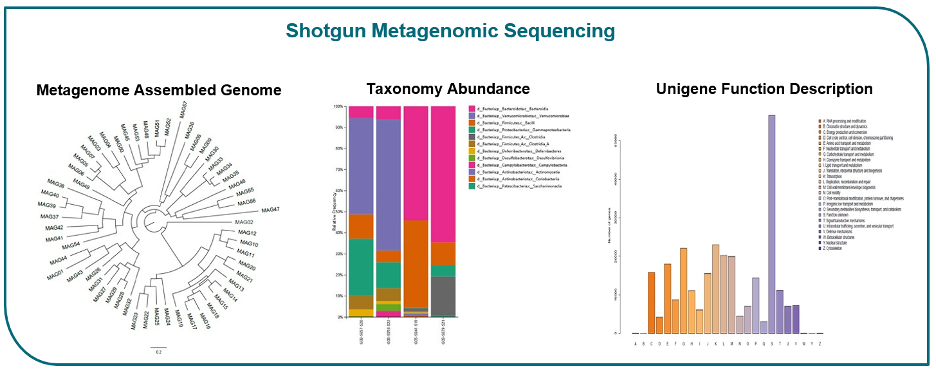
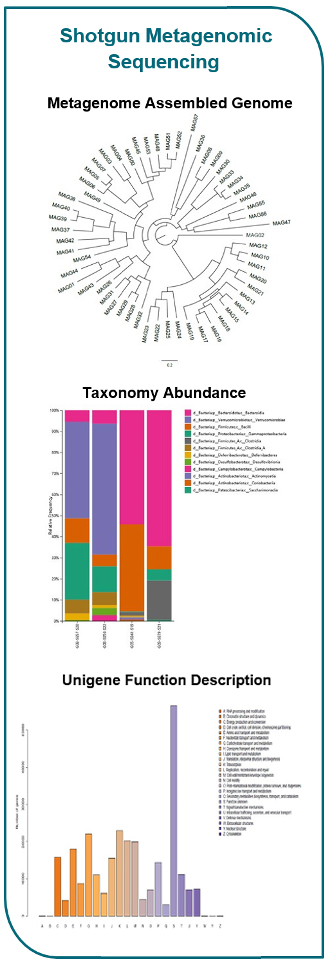
Metatranscriptomic sequencing studies the gene expression profiles of microbial communities, providing insights into their functional activities and responses to the environment. By analyzing RNA molecules, researchers can identify actively expressed genes, revealing the metabolic pathways and biological processes within the microbiome. This approach enhances our understanding of how microbes interact with their surroundings and contribute to ecosystem functions and host-associated processes.
|
Methods |
Applications |
Advantages |
Limitations |
|
Full-length 16S rRNA |
|
|
|
|
Shotgun Metagenomic |
|
|
|
|
Metatranscriptomic |
|
|
|
© 2024 Crown Bioscience. All Rights Reserved.


© 2024 Crown Bioscience. All Rights Reserved. Privacy Policy
2023-11-29
2023-10-20
site_page
Genomics