- Our Services
- Platforms
- Target Solutions
- Technologies
- Service Types
- Our Science
- About Us
- Contact us
Yanghui Sheng, Wubin Qian, Xiaobo Chen, Henry Q. Li, Sheng Guo
Crown Bioscience, Inc., 218 Xinghu Street, Suzhou, Jiangsu, 215000, China 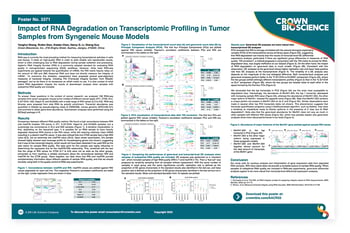 RNA-seq is currently the most prevailing method for measuring transcriptional activities in cells and tissues. It relies on high-quality RNA in order to yield reliable and reproducible results, which is often challenging due to RNA degradation during sample collection and processing. Agilent’s RNA Integrity Number (RIN) is a commonly adopted standard for evaluating RNA quality in next-generation sequencing (NGS) workflows.
RNA-seq is currently the most prevailing method for measuring transcriptional activities in cells and tissues. It relies on high-quality RNA in order to yield reliable and reproducible results, which is often challenging due to RNA degradation during sample collection and processing. Agilent’s RNA Integrity Number (RIN) is a commonly adopted standard for evaluating RNA quality in next-generation sequencing (NGS) workflows.
However, while most RNA-seq experiments are geared towards the quantification of mRNA, the RIN metric heavily relies on the amount of 18S and 28S ribosomal RNA and does not directly measure the integrity of mRNA1. To overcome this limitation, researchers have proposed several post-alignment measures of transcript integrity, including TIN (Transcript Integrity Number, from RSeQC package)2, but so far there is no consensus on which metric to use. It is also unclear to what extent RNA degradation impacts the results of downstream analysis when samples with suboptimal RNA quality are included.
Your privacy is important to us.
We'll never share your information.
© 2025 Crown Bioscience. All Rights Reserved.
Privacy Policy | Imprint | Terms of Service | Privacy Preferences


© 2025 Crown Bioscience. All Rights Reserved. Privacy Policy
2023-05-22
2022-04-06
landing_page
PDX/Databases